TheorChromo is an online tool for peptide and protein retention time prediction in liquid chromatography. It implements the BioLCCC model for the most popular case of linear gradient chromatography.
Just follow the instructions on the main page.
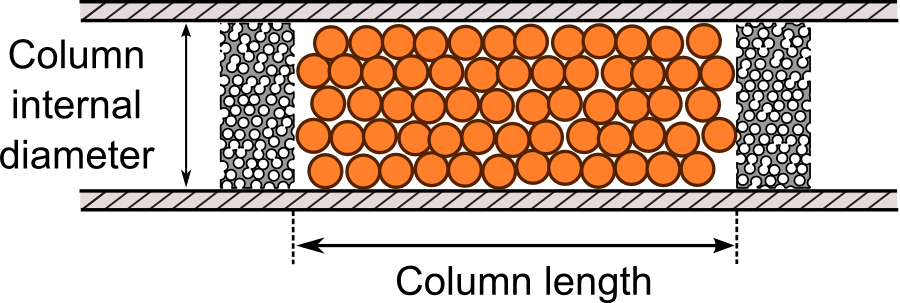
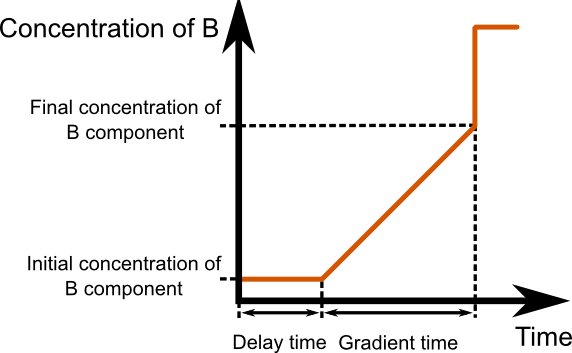
Firstly, enter the parameters of your LC
system in the corresponding lines.
WARNING: All parameters are saved between sessions.
RP/ACN+FA - Magic C18 AQ reversed solid phase, ACN as an eluent and 0.1% formic acid in both solvents;
RP/ACN+TFA - SynChropak RP-P C18 column, ACN as an eluent and 0.1% trifluoroacetic acid in both solvents.
On the next step you need to enter peptide sequences.
The sequences should be written using the standard one-letter codes
for the twenty proteinogenic L-amino acids and the extended notation
for modified amino acids.
WARNING: Only capital letters are allowed for the standard amino
acids.
WARNING: Cysteine is assumed to be pure! If the peptides were treated
with iodoacetamide, check the box "Cysteines are
carboxyamidomethylated" or use the corresponding notation.
TheorChromo is based on the BioLCCC (Liquid Chromatography of Biomacromolecules at Critical Conditions) model. Contrary to the other models of peptide chromatography, BioLCCC uses only very basic assumptions regarding flexibility of a polypeptide chain, the shape of a pore, type of interactions neglected, etc. Given this assumptions, the coefficient of distribution (Kd) of a peptide between solid phase and mobile phase of a given composition is derived using the methods of statistical physics of macromolecules. Finally, the retention time of a peptide is derived from Kd using the basis equation of gradient chromatography. TheorChromo is based on the BioLCCC (Liquid Chromatography of Biomolecules at Critical Conditions) model which describes the adsorption of protein molecules on porous media. Contrary to the other models of peptide/protein chromatography, BioLCCC starts from very basic assumptions regarding flexibility of a polypeptide chain, the shape of a pore, type of interactions neglected, etc. Given this assumptions, the coefficient of distribution (Kd) of a peptide between solid phase and mobile phase can be derived using the methods of statistical physics of macromolecules. Finally, the retention time of a peptide is calculated from Kd using the basis equation of gradient chromatography.
Owing to the physical basis of the BioLCCC model, it contains very few free parameters. The retention properties of an amino acid are characterized by a single number, which is essentially the energy of interaction between the amino acid and the surface of solid phase in pure water+ion paring agent. Given this small number of phenomenological parameters, the BioLCCC model can easily be adapted for an arbitrary type of chromatography not limited by phase or solvent types. Moreover, its extension to peptides with post-translational modifications is straightforward as it was shown for the phosphorylated amino acids.
Further reading:
1. Liquid Chromatography at Critical Conditions: Comprehensive
Approach to Sequence-Dependent Retention Time Prediction,
Alexander V. Gorshkov, Irina A. Tarasova, Victor V. Evreinov, Mikhail M. Savitski, Michael L. Nielsen, Roman A. Zubarev, and Mikhail V. Gorshkov,
Analytical Chemistry, 2006, 78 (22), 7770-7777.
2. Applicability of the critical chromatography concept to proteomics
problems: Dependence of retention time on the sequence of amino
acids, Alexander V. Gorshkov A., Victor V. Evreinov V., Irina A.
Tarasova, Mikhail V. Gorshkov, Polymer Science B, 2007,
49 (3-4), 93-107.
3. Applicability of the critical chromatography concept to proteomics
problems: Experimental study of the dependence of peptide retention time
on the sequence of amino acids in the chain,
Irina A. Tarasova, Alexander V. Gorshkov, Victor V. Evreinov,
Chris Adams, Roman A. Zubarev, and Mikhail V. Gorshkov,
Polymer Science A, 2008, 50 (3), 309.
Yes, the source code of BioLCCC is open and can be used in any free project. Please, check libBioLCCC and, if you use Python in your work, pyteomics.biolccc projects.
You are welcome to the BioLCCC discussion group: http://groups.google.com/group/biolccc